Integrating into Pipelines for Clustering¶
Starting off, we import a few required Python resources. While there are quite a few in there, notice we are grabbing the make_pipeline method from Sci-Kit Learn. We are going to be building pipelines!
[1]:
import os
from pathlib import Path
import sys
from arcgis.features import GeoAccessor
from arcgis.geoenrichment import Country
from arcgis.gis import GIS
from dotenv import find_dotenv, load_dotenv
from matplotlib import pyplot as plt
import numpy as np
import pandas as pd
from sklearn.pipeline import make_pipeline
# load the "autoreload" extension so that code can change, & always reload modules so that as you change code in src, it gets loaded
%load_ext autoreload
%autoreload 2
[2]:
# load environment variables from .env
load_dotenv(find_dotenv())
# paths to input data
dir_prj = Path.cwd().parent
dir_data = dir_prj/'data'
dir_raw = dir_data/'raw'
# import the two preprocessors from the examples
sys.path.append(str(dir_prj/'src'))
from ba_samples.preprocessing import EnrichStandardGeography, KeepOnlyEnrichColumns, ArrayToDataFrame
# specifically, the data being used for this example - pickled dataframes
postal_codes_pth = dir_raw/'pdx_postal_codes.pkl'
block_groups_pth = dir_raw/'pdx_block_groups.pkl'
Pipeline for Zip Codes¶
We start by getting a list of standard geography codes to use from the demonstration postal code data, just a list of zip codes.
[3]:
postal_code_df = pd.read_pickle(postal_codes_pth)
postal_code_lst = list(postal_code_df['ID'])
print(postal_code_lst)
['97038', '97101', '97304', '97347', '97362', '97375', '97396', '97002', '97003', '97004', '97005', '97006', '97007', '97008', '97009', '97013', '97015', '97016', '97017', '97018', '97022', '97024', '97027', '97030', '97032', '97034', '97035', '97036', '97042', '97045', '97048', '97051', '97053', '97054', '97056', '97060', '97062', '97064', '97068', '97070', '97071', '97078', '97080', '97086', '97089', '97106', '97109', '97111', '97113', '97114', '97115', '97116', '97117', '97119', '97123', '97124', '97125', '97127', '97128', '97132', '97133', '97140', '97144', '97148', '97201', '97202', '97203', '97204', '97205', '97206', '97209', '97210', '97211', '97212', '97213', '97214', '97215', '97216', '97217', '97218', '97219', '97220', '97221', '97222', '97223', '97224', '97225', '97227', '97229', '97230', '97231', '97232', '97233', '97236', '97239', '97266', '97267', '97378', '98601', '98604', '98606', '98607', '98629', '98642', '98649', '98660', '98661', '98662', '98663', '98664', '98665', '98674', '98675', '98682', '98683', '98684', '98685', '98686', '97023', '97010', '97011', '97014', '97019', '97028', '97049', '97055', '97067', '98605', '98610', '98639', '98648', '98651', '98671']
Get a Local Country¶
To enrich, we need a Country object instance. As part of the constructor, we need to tell the object what Business Analyst source to use in the gis parameter. In this case, we are telling the object to use ArcGIS Pro with Business Analyst and local data for the United States.
[4]:
usa_local = Country('usa', gis=GIS('pro'))
usa_local
[4]:
<Country - United States 2021 ('local')>
Select Variables¶
Next, we need to get some enrich variables to use. We can discover what is available using the enrich_variables property of the country object to retrieve a Pandas Data Frame of variables available for the country. From these tens of thousands of variables, we can prune this down to a manageable subset.
[5]:
ev = usa_local.enrich_variables
kv = ev[
(ev.data_collection.str.lower().str.contains('key'))
& (ev.name.str.lower().str.endswith('cy'))
].reset_index(drop=True)
kv
[5]:
| name | alias | data_collection | enrich_name | enrich_field_name | |
|---|---|---|---|---|---|
| 0 | TOTPOP_CY | 2021 Total Population | KeyUSFacts | KeyUSFacts.TOTPOP_CY | KeyUSFacts_TOTPOP_CY |
| 1 | GQPOP_CY | 2021 Group Quarters Population | KeyUSFacts | KeyUSFacts.GQPOP_CY | KeyUSFacts_GQPOP_CY |
| 2 | DIVINDX_CY | 2021 Diversity Index | KeyUSFacts | KeyUSFacts.DIVINDX_CY | KeyUSFacts_DIVINDX_CY |
| 3 | TOTHH_CY | 2021 Total Households | KeyUSFacts | KeyUSFacts.TOTHH_CY | KeyUSFacts_TOTHH_CY |
| 4 | AVGHHSZ_CY | 2021 Average Household Size | KeyUSFacts | KeyUSFacts.AVGHHSZ_CY | KeyUSFacts_AVGHHSZ_CY |
| 5 | MEDHINC_CY | 2021 Median Household Income | KeyUSFacts | KeyUSFacts.MEDHINC_CY | KeyUSFacts_MEDHINC_CY |
| 6 | AVGHINC_CY | 2021 Average Household Income | KeyUSFacts | KeyUSFacts.AVGHINC_CY | KeyUSFacts_AVGHINC_CY |
| 7 | PCI_CY | 2021 Per Capita Income | KeyUSFacts | KeyUSFacts.PCI_CY | KeyUSFacts_PCI_CY |
| 8 | TOTHU_CY | 2021 Total Housing Units | KeyUSFacts | KeyUSFacts.TOTHU_CY | KeyUSFacts_TOTHU_CY |
| 9 | OWNER_CY | 2021 Owner Occupied HUs | KeyUSFacts | KeyUSFacts.OWNER_CY | KeyUSFacts_OWNER_CY |
| 10 | RENTER_CY | 2021 Renter Occupied HUs | KeyUSFacts | KeyUSFacts.RENTER_CY | KeyUSFacts_RENTER_CY |
| 11 | VACANT_CY | 2021 Vacant Housing Units | KeyUSFacts | KeyUSFacts.VACANT_CY | KeyUSFacts_VACANT_CY |
| 12 | MEDVAL_CY | 2021 Median Home Value | KeyUSFacts | KeyUSFacts.MEDVAL_CY | KeyUSFacts_MEDVAL_CY |
| 13 | AVGVAL_CY | 2021 Average Home Value | KeyUSFacts | KeyUSFacts.AVGVAL_CY | KeyUSFacts_AVGVAL_CY |
| 14 | POPGRW10CY | 2010-2021 Growth Rate: Population | KeyUSFacts | KeyUSFacts.POPGRW10CY | KeyUSFacts_POPGRW10CY |
| 15 | HHGRW10CY | 2010-2021 Growth Rate: Households | KeyUSFacts | KeyUSFacts.HHGRW10CY | KeyUSFacts_HHGRW10CY |
| 16 | FAMGRW10CY | 2010-2021 Growth Rate: Families | KeyUSFacts | KeyUSFacts.FAMGRW10CY | KeyUSFacts_FAMGRW10CY |
| 17 | DPOP_CY | 2021 Total Daytime Population | KeyUSFacts | KeyUSFacts.DPOP_CY | KeyUSFacts_DPOP_CY |
| 18 | DPOPWRK_CY | 2021 Daytime Pop: Workers | KeyUSFacts | KeyUSFacts.DPOPWRK_CY | KeyUSFacts_DPOPWRK_CY |
| 19 | DPOPRES_CY | 2021 Daytime Pop: Residents | KeyUSFacts | KeyUSFacts.DPOPRES_CY | KeyUSFacts_DPOPRES_CY |
Get Standard Geography Name¶
Using the levels property, we can see the value we need to use for specifying the specfic zip code level, zip5.
[6]:
usa_local.levels
[6]:
| level_name | alias | level_id | id_field | name_field | singular_name | plural_name | admin_level | |
|---|---|---|---|---|---|---|---|---|
| 0 | block_groups | Block Groups | US.BlockGroups | ID | NAME | Block Group | Block Groups | Admin11 |
| 1 | tracts | Census Tracts | US.Tracts | ID | NAME | Census Tract | Census Tracts | Admin10 |
| 2 | places | Cities and Towns (Places) | US.Places | ID | NAME | Place | Places | Admin9 |
| 3 | zip5 | ZIP Codes | US.ZIP5 | ID | NAME | ZIP Code | ZIP Codes | Admin4 |
| 4 | csd | County Subdivisions | US.CSD | ID | NAME | County Subdivision | County Subdivisions | Admin7 |
| 5 | counties | Counties | US.Counties | ID | NAME | County | Counties | Admin3 |
| 6 | cbsa | CBSAs | US.CBSA | ID | NAME | CBSA | CBSAs | Admin5 |
| 7 | cd | Congressional Districts | US.CD | ID | NAME | Congressional District | Congressional Districts | Admin8 |
| 8 | dma | DMAs | US.DMA | ID | NAME | DMA | DMAs | Admin6 |
| 9 | states | States | US.States | ID | NAME | State | States | Admin2 |
| 10 | whole_usa | Entire Country | US.WholeUSA | ID | NAME | United States of America | United States of America | Admin1 |
Enrich Preprocessor¶
The enrich method can be wrapped into the transform method of a SciKit-Learn Transformer, specifically a preprocessor, to function just like any other preprocessor. In this case, not only have I wrapped the preprocessor, I also have created this preprocessor for specific inputs, standard geographies.
[7]:
from ba_samples.preprocessing import EnrichStandardGeography
enrich_preprocessor = EnrichStandardGeography(usa_local, enrich_variables=kv, standard_geography_level='zip5', return_geometry=False)
enrich_df = enrich_preprocessor.fit_transform(postal_code_lst)
enrich_df.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 133 entries, 0 to 132
Data columns (total 26 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 id_field 133 non-null object
1 area_desc 133 non-null object
2 ta_desc 133 non-null object
3 names 133 non-null object
4 has_data 133 non-null int32
5 aggregation_method 133 non-null object
6 totpop_cy 133 non-null float64
7 gqpop_cy 133 non-null float64
8 divindx_cy 133 non-null float64
9 tothh_cy 133 non-null float64
10 avghhsz_cy 133 non-null float64
11 medhinc_cy 133 non-null float64
12 avghinc_cy 133 non-null float64
13 pci_cy 133 non-null float64
14 tothu_cy 133 non-null float64
15 owner_cy 133 non-null float64
16 renter_cy 133 non-null float64
17 vacant_cy 133 non-null float64
18 medval_cy 133 non-null float64
19 avgval_cy 133 non-null float64
20 popgrw10_cy 133 non-null float64
21 hhgrw10_cy 133 non-null float64
22 famgrw10_cy 133 non-null float64
23 dpop_cy 133 non-null float64
24 dpopwrk_cy 133 non-null float64
25 dpopres_cy 133 non-null float64
dtypes: float64(20), int32(1), object(5)
memory usage: 26.6+ KB
[8]:
enrich_df.head()
[8]:
| id_field | area_desc | ta_desc | names | has_data | aggregation_method | totpop_cy | gqpop_cy | divindx_cy | tothh_cy | ... | renter_cy | vacant_cy | medval_cy | avgval_cy | popgrw10_cy | hhgrw10_cy | famgrw10_cy | dpop_cy | dpopwrk_cy | dpopres_cy | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 97038 | 97038 | TA from geography Layer: 97038 | Molalla | 1 | BlockApportionment:US.BlockGroups;PointsLayer:... | 17305.0 | 56.0 | 42.4 | 6144.0 | ... | 1552.0 | 293.0 | 388300.0 | 442226.0 | 1.18 | 1.20 | 1.00 | 13803.0 | 4468.0 | 9335.0 |
| 1 | 97002 | 97002 | TA from geography Layer: 97002 | Aurora | 1 | BlockApportionment:US.BlockGroups;PointsLayer:... | 5951.0 | 52.0 | 54.3 | 2285.0 | ... | 389.0 | 85.0 | 491246.0 | 565932.0 | 1.09 | 1.15 | 0.97 | 7040.0 | 3847.0 | 3193.0 |
| 2 | 97347 | 97347 | TA from geography Layer: 97347 | Grand Ronde | 1 | BlockApportionment:US.BlockGroups;PointsLayer:... | 2163.0 | 0.0 | 53.5 | 850.0 | ... | 242.0 | 60.0 | 375287.0 | 446223.0 | 1.28 | 1.31 | 1.13 | 2296.0 | 907.0 | 1389.0 |
| 3 | 97013 | 97013 | TA from geography Layer: 97013 | Canby | 1 | BlockApportionment:US.BlockGroups;PointsLayer:... | 25403.0 | 104.0 | 54.2 | 9139.0 | ... | 2458.0 | 340.0 | 392366.0 | 437940.0 | 0.96 | 0.98 | 0.85 | 21914.0 | 9136.0 | 12778.0 |
| 4 | 97017 | 97017 | TA from geography Layer: 97017 | Colton | 1 | BlockApportionment:US.BlockGroups;PointsLayer:... | 3194.0 | 0.0 | 16.9 | 1133.0 | ... | 125.0 | 58.0 | 434483.0 | 480442.0 | 0.68 | 0.72 | 0.58 | 2405.0 | 687.0 | 1718.0 |
5 rows × 26 columns
Filtering Columns¶
The output from enrich includes a number of metadata columns. If performing subsequent analysis, we need to prune these columns. We can use another preprocessor for this as well, one only keeping columns with enriched values.
[9]:
from ba_samples.preprocessing import KeepOnlyEnrichColumns
enrich_zip_pipe = make_pipeline(
EnrichStandardGeography(usa_local, enrich_variables=kv, standard_geography_level='zip5', return_geometry=False),
KeepOnlyEnrichColumns(usa_local, id_column='id_field', keep_geometry=False)
)
enrich_df = enrich_zip_pipe.fit_transform(postal_code_lst)
enrich_df.info()
<class 'pandas.core.frame.DataFrame'>
Index: 133 entries, 97038 to 98671
Data columns (total 17 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 totpop_cy 133 non-null float64
1 gqpop_cy 133 non-null float64
2 divindx_cy 133 non-null float64
3 tothh_cy 133 non-null float64
4 avghhsz_cy 133 non-null float64
5 medhinc_cy 133 non-null float64
6 avghinc_cy 133 non-null float64
7 pci_cy 133 non-null float64
8 tothu_cy 133 non-null float64
9 owner_cy 133 non-null float64
10 renter_cy 133 non-null float64
11 vacant_cy 133 non-null float64
12 medval_cy 133 non-null float64
13 avgval_cy 133 non-null float64
14 dpop_cy 133 non-null float64
15 dpopwrk_cy 133 non-null float64
16 dpopres_cy 133 non-null float64
dtypes: float64(17)
memory usage: 18.7+ KB
[10]:
enrich_df.head()
[10]:
| keyusfacts_totpop_cy | keyusfacts_gqpop_cy | keyusfacts_divindx_cy | keyusfacts_tothh_cy | keyusfacts_avghhsz_cy | keyusfacts_medhinc_cy | keyusfacts_avghinc_cy | keyusfacts_pci_cy | keyusfacts_tothu_cy | keyusfacts_owner_cy | keyusfacts_renter_cy | keyusfacts_vacant_cy | keyusfacts_medval_cy | keyusfacts_avgval_cy | keyusfacts_popgrw10cy | keyusfacts_hhgrw10cy | keyusfacts_famgrw10cy | keyusfacts_dpop_cy | keyusfacts_dpopwrk_cy | keyusfacts_dpopres_cy | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id_field | ||||||||||||||||||||
| 83801 | 8497.0 | 0.0 | 13.6 | 3142.0 | 2.70 | 62413.0 | 78746.0 | 30156.0 | 3563.0 | 2660.0 | 482.0 | 421.0 | 382542.0 | 391184.0 | 2.07 | 2.09 | 1.92 | 6502.0 | 1962.0 | 4540.0 |
| 83803 | 656.0 | 0.0 | 10.5 | 341.0 | 1.92 | 43344.0 | 61732.0 | 29472.0 | 659.0 | 264.0 | 76.0 | 318.0 | 391333.0 | 403113.0 | 1.76 | 1.80 | 1.56 | 499.0 | 145.0 | 354.0 |
| 83810 | 1090.0 | 4.0 | 11.7 | 479.0 | 2.27 | 60088.0 | 89006.0 | 38340.0 | 579.0 | 406.0 | 72.0 | 100.0 | 437903.0 | 464383.0 | 0.94 | 0.96 | 0.76 | 762.0 | 246.0 | 516.0 |
| 83814 | 28546.0 | 609.0 | 21.2 | 12454.0 | 2.24 | 57487.0 | 80524.0 | 34960.0 | 15054.0 | 6855.0 | 5599.0 | 2600.0 | 377722.0 | 508589.0 | 1.86 | 1.91 | 1.72 | 38178.0 | 23011.0 | 15167.0 |
| 83815 | 38614.0 | 617.0 | 24.1 | 15247.0 | 2.49 | 56872.0 | 73431.0 | 29323.0 | 16393.0 | 9649.0 | 5598.0 | 1146.0 | 308198.0 | 374194.0 | 1.79 | 1.78 | 1.51 | 37345.0 | 15891.0 | 21454.0 |
Clustering Block Groups Using Demographics¶
Stepping up our game a little, now we are getting all the US Census Block Groups in Portland because we are interested in what demographic variables have the most differences in Portland, and are going to use a SciKit-Learn pipeline to streamline the process.
[10]:
block_groups_df = pd.read_pickle(block_groups_pth)
block_groups_lst = list(block_groups_df['ID'])
print(f'Block Group Count: {len(block_groups_lst):,}')
Block Group Count: 1,421
Source - ArcGIS Online¶
This time around, we no longer are using local resources. Now, we are switching to ArcGIS Online.
[12]:
gis_agol = GIS(url=os.getenv('ESRI_GIS_URL'), username=os.getenv('ESRI_GIS_USERNAME'), password=os.getenv('ESRI_GIS_PASSWORD'))
usa_agol = Country('usa', gis=gis_agol)
usa_agol
[12]:
<Country - United States (GIS @ https://baqa.mapsqa.arcgis.com version:9.4)>
Selecting Variables¶
This time around, we are going big. By just grabbing a lot of categories, we are selecting a wide swath of demographic varaiables. There wasn not a tremenedously emperical method used to retrieve these categories. I just grabbed quite a few. Below, please notice how I used some naming conventions to filter out some values to exclude.
[13]:
collections = ['classofworker', 'commute', 'disability', 'disposableincome', 'disposableincome', 'educationalattainment',
'financial', 'foodstampssnap', 'gender', 'generations', 'groupquarters', 'health', 'healthinsurancecoverage',
'homevalue', 'householdincome', 'households', 'householdsbyageofhouseholder', 'householdsbysize',
'householdtotals', 'householdtype', 'housingbyageofhouseholder', 'housingbysize', 'housingcosts',
'housinghousehold', 'housingunittotals', 'industry', 'internetcomputerusage', 'keyusfacts',
'language', 'network', 'occupation', 'wealth', 'veterans']
ev = usa_agol.enrich_variables
enrich_vars = ev[
(ev.data_collection.isin(collections))
& (~ev.alias.str.lower().str.contains('index')) # exclude calculated indexes
& (~ev.name.str.lower().str.endswith('_a')) # exclude averages
& (~ev.name.str.lower().str.contains('_fy')) # exclude future year
& (~ev.name.str.lower().str.contains('fy_')) # exclude another future year pattern
& (~ev.alias.str.contains('2010')) # exclude 2010 census variables - we're not living in the past anymore
].drop_duplicates('name').reset_index(drop=True)
enrich_vars.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 1686 entries, 0 to 1685
Data columns (total 8 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 name 1686 non-null object
1 alias 1686 non-null object
2 data_collection 1686 non-null object
3 enrich_name 1686 non-null object
4 enrich_field_name 1686 non-null object
5 description 1686 non-null object
6 vintage 1686 non-null object
7 units 1686 non-null object
dtypes: object(8)
memory usage: 105.5+ KB
Just double checking, we can quickly see the correct value we need to use for United States Census Block Groups is block_groups.
[14]:
usa_agol.levels
[14]:
| level_name | singular_name | plural_name | alias | level_id | admin_level | |
|---|---|---|---|---|---|---|
| 0 | block_groups | Block Group | Block Groups | Block Groups | US.BlockGroups | |
| 1 | tracts | Census Tract | Census Tracts | Census Tracts | US.Tracts | |
| 2 | places | Place | Places | Cities and Towns (Places) | US.Places | |
| 3 | zip5 | ZIP Code | ZIP Codes | ZIP Codes | US.ZIP5 | Admin4 |
| 4 | csd | County Subdivision | County Subdivisions | County Subdivisions | US.CSD | |
| 5 | counties | County | Counties | Counties | US.Counties | Admin3 |
| 6 | cbsa | CBSA | CBSAs | CBSAs | US.CBSA | |
| 7 | cd | Congressional District | Congressional Districts | Congressional Districts | US.CD | |
| 8 | dma | DMA | DMAs | DMAs | US.DMA | |
| 9 | states | State | States | States | US.States | Admin2 |
| 10 | whole_usa | United States of America | United States of America | Entire Country | US.WholeUSA | Admin1 |
Get Demographics¶
Just like before, we are going to enrich, but this time around we are using a LOT of data - over 1,500 variables. This time around, we are including the geometry so we can use it later.
[15]:
# run the enrich steps autonomously so we can access the keeper properties in later steps
enrich_pipe = make_pipeline(
EnrichStandardGeography(usa_agol, enrich_variables=enrich_vars, standard_geography_level='block_groups', return_geometry=True),
KeepOnlyEnrichColumns(usa_agol, keep_geometry=True)
)
enrich_df = enrich_pipe.fit_transform(block_groups_lst)
enrich_df.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 1421 entries, 0 to 1420
Columns: 1324 entries, acscivemp to SHAPE
dtypes: float64(67), geometry(1), int64(1256)
memory usage: 14.4 MB
Get Just the Enriched Columns¶
Following enrichment, now we are getting just the columns not containing the geometry, the polygon shapes of the Block Groups. Notice the geometry date type is no longer listed.
[16]:
demographic_cols = [c for c in enrich_df.columns if c != enrich_df.spatial.name]
training_df = enrich_df.loc[:,demographic_cols]
training_df.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 1421 entries, 0 to 1420
Columns: 1323 entries, acscivemp to acstotpop
dtypes: float64(67), int64(1256)
memory usage: 14.3 MB
Data Cleanup¶
Since subsequent steps need standard scaled data and have difficulty with zero variance and missing values, we use tools from Sci-Kit Learn to clean up the data.
[17]:
from sklearn.cluster import KMeans
from sklearn.feature_selection import VarianceThreshold
from sklearn.impute import SimpleImputer
from sklearn.preprocessing import MinMaxScaler
scale_pipe = make_pipeline(
MinMaxScaler(), # get on same scale - just using because it is simple
VarianceThreshold(threshold=0.0), # drop zero variance variables
SimpleImputer() # using defaults, filling nulls with mean
)
scaled_arr = scale_pipe.fit_transform(training_df)
scaled_arr
[17]:
array([[0.11816762, 0.16923077, 0.66666667, ..., 0. , 0. ,
0.19456258],
[0.1832379 , 0.17342657, 0.33333333, ..., 0. , 0. ,
0.23329283],
[0.07105674, 0.18881119, 0.66666667, ..., 0. , 0. ,
0.1240887 ],
...,
[0.15955232, 0.12167832, 0.33333333, ..., 0. , 0. ,
0.20990279],
[0.0840708 , 0.23636364, 0.66666667, ..., 0. , 0. ,
0.13456865],
[0.26106195, 0.21818182, 0.33333333, ..., 0. , 0. ,
0.30726002]])
Dimensionalty Reduction¶
Over 1,000 variables is a lot to process. To speed up the process, we first are going to use Principal Component Analysis (PCA) for dimensionality reduction, using a scree plot to ascertain how many components to create. In this case, we are going with 200 to retain roughly 90% of the variation in the data.
[18]:
from sklearn.decomposition import PCA
pca = PCA().fit(scaled_arr)
plt.style.use('ggplot')
plt.figure(figsize=(10,8))
plt.plot(np.cumsum(pca.explained_variance_ratio_))
plt.xlabel('number of components')
plt.ylabel('cumulative explained variance')
plt.yticks([0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 1.0])
plt.show()
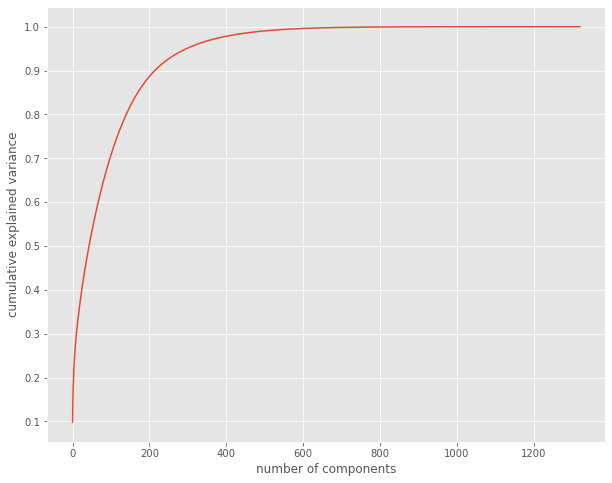
Clustering¶
From the scree plot we know the number of clusters to use with PCA, so we are now going to combine this with K-Means Clustering in a succinct pipeline. Once we have the clusters created, we then will create a new spatially enabled Pandas data frame by combining the output clusters with the geometry returned from geoenrichment earlier.
[19]:
from sklearn.cluster import KMeans
cluster_pipe = make_pipeline(
PCA(n_components=200),
KMeans(n_clusters=5)
)
cluster_pipe.fit_transform(scaled_arr)
cluster_df = pd.DataFrame(zip(block_groups_lst, cluster_pipe.named_steps.kmeans.labels_, enrich_df['SHAPE']),
columns=['fips', 'cluster_id', 'SHAPE'])
cluster_df.spatial.set_geometry('SHAPE')
cluster_df.info()
cluster_df.head()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 1421 entries, 0 to 1420
Data columns (total 3 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 fips 1421 non-null object
1 cluster_id 1421 non-null int32
2 SHAPE 1421 non-null geometry
dtypes: geometry(1), int32(1), object(1)
memory usage: 27.9+ KB
[19]:
| fips | cluster_id | SHAPE | |
|---|---|---|---|
| 0 | 410710305021 | 2 | {"rings": [[[-123.56601700006303, 45.216389999... |
| 1 | 410050237001 | 1 | {"rings": [[[-122.61617099963205, 45.267457999... |
| 2 | 410050237002 | 4 | {"rings": [[[-122.5704800001157, 45.2375169997... |
| 3 | 410050237003 | 1 | {"rings": [[[-122.5111050007072, 45.2601139996... |
| 4 | 410050237004 | 1 | {"rings": [[[-122.50749899966338, 45.230098999... |
Dissolve Clusters¶
For visualization on a map and also for further investigation using Inforgaphics, we need the geometries consolidated into a single geometry per cluster. The ArcGIS Online Geometry engine can be used through the arcgis.geometry.union method to accomplish this.
[20]:
from arcgis.geometry import union
def dissolve_by_cluster_id(cluster_id):
"""Helper to dissolve geometries based on the cluster_id."""
# pull all the geometries out of the cluster dataframe matching the cluster id as a list
cluster_geom_lst = list(cluster_df[cluster_df['cluster_id'] == cluster_id]['SHAPE'])
# use the ArcGIS Online geometry service to combine all the geometeries into one
dissolved_cluster_geom = union(cluster_geom_lst, gis=gis_agol)
return dissolved_cluster_geom
[21]:
uniq_cluster_id_lst = cluster_df['cluster_id'].unique()
uniq_cluster_id_lst.sort()
cluster_geom_lst = []
for c_id in uniq_cluster_id_lst:
c_geom = dissolve_by_cluster_id(c_id)
cluster_geom_lst.append(c_geom)
print(f'Created geometry for cluster id {c_id}')
dissolved_cluster_df = pd.DataFrame(zip(uniq_cluster_id_lst, cluster_geom_lst), columns=['cluster_id', 'SHAPE'])
dissolved_cluster_df.spatial.set_geometry('SHAPE')
dissolved_cluster_df
Created geometry for cluster id 0
Created geometry for cluster id 1
Created geometry for cluster id 2
Created geometry for cluster id 3
Created geometry for cluster id 4
[21]:
| cluster_id | SHAPE | |
|---|---|---|
| 0 | 0 | {"rings": [[[-122.58948899999996, 45.181993000... |
| 1 | 1 | {"rings": [[[-122.61984599899995, 45.118054999... |
| 2 | 2 | {"rings": [[[-123.38401899999997, 45.110430000... |
| 3 | 3 | {"rings": [[[-123.45598799999999, 45.125803999... |
| 4 | 4 | {"rings": [[[-123.37559099899994, 45.094734000... |
Visualizing, Sharing and Interrogating¶
Both outputs from the clustering are spatially enabled Pandas data frames, and can easily be shared with ArcGIS Online. From there, the results of these analyses can be quickly visualized in maps and mapping applications, shared to communicate results, and further interrogated through Infographics.
[22]:
bg_lyr = cluster_df.spatial.to_featurelayer('pdx_cluster_block_groups',
gis=gis_agol,
tags=['pdx', 'machine learning', 'clustering'],
service_name='pdx_cluster_block_groups')
bg_lyr
[22]:
[23]:
clstr_lyr = dissolved_cluster_df.spatial.to_featurelayer('pdx_clusters',
gis=gis_agol,
tags=['pdx', 'machine learning', 'clustering'],
service_name='pdx_clusters')
clstr_lyr
[23]:
[ ]:
